GO term network
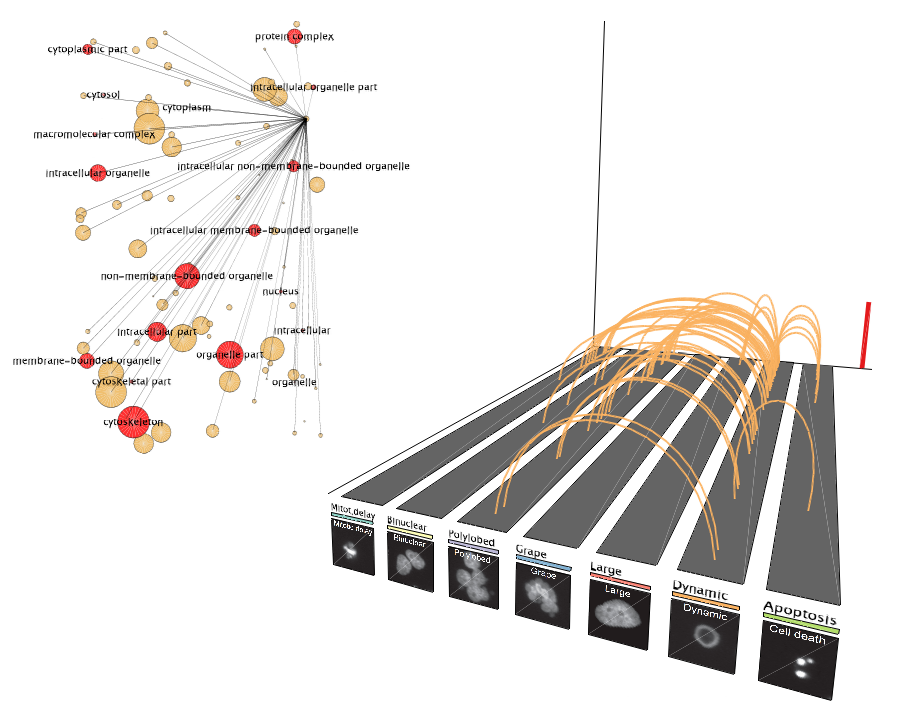
The Gene Ontology (GO) term network can be displayed in parallel with the phenotypic connections. This can provide some insight into what kind of functions the genes underlying the connections have. The network changes dynamically at each time point by highlighting (in red) only those terms that correspond to the genes responsible for the connections at the particular moment in time.
The size of the nodes is proportional to the number of enriched genes for that function in the entire dataset.
The user can change between 3 different GO representations, if available: biological process, molecular function and cellular component.
The GO term network is layed out using the force-directed algorithm rather than a hierarchical one for space optimization constraints. Hierarchical layout is however planned for a future release. The network is interactive: nodes can be moved around and clicking on a node highlights its links to other nodes.
For the moment, the GO network cannot be automatically generated, it has to be loaded by the user.
